Accelerate your protein engineering. Download our free guide to cell display
Troubleshooting Low Display Levels in Yeast and Mammalian Cells: A Step-by-Step Checklist
Low or non-existent display levels are a common roadblock that can halt a yeast display or mammalian display campaign. This guide provides a step-by-step checklist to systematically diagnose the root cause, whether it's a flawed protocol, an incorrect plasmid construct, or a sign of poor developability. By following this framework, researchers can not only troubleshoot their experiments but also leverage low display as a valuable filter to select for more stable and well-behaved protein candidates.
10/2/20254 min read
No fluorescence signal on your flow cytometer is never a good start to any yeast display or mammalian display campaign. After weeks of library construction, transformations and careful preparation, you stain for your expression tag (like c-myc or HA) only to find that the fluorescence signal is indistinguishable from the negative control. Low or non-existent display is a common roadblock that can bring a project to a screeching halt. The cause can range from a trivial protocol error to a fundamental issue with the protein variant itself.
Here are some common things to look for when troubleshooting low display levels, whether you are working with a full library or individual clones.
The First Diagnostic: Is the Problem Global or Clone-Specific?
Before you begin, you must answer one critical question: Is the entire library showing low display, or is the issue confined to a specific subset of clones?
Global Problem: If your unselected library and all your controls (including a known positive control binder) show low display, the issue likely lies with your core system: the vector, the host cells, or the experimental protocol.
Clone-Specific Problem: If your positive control looks fine but certain clones or enriched pools show progressively lower display levels after each round of selection, the issue is almost certainly with the protein variants themselves.
Your answer to this question will determine which troubleshooting path to follow.
Path 1: Troubleshooting Global Low Display (System-Wide Issues)
If your entire population is failing to display properly, start by investigating the foundational components of your experiment.
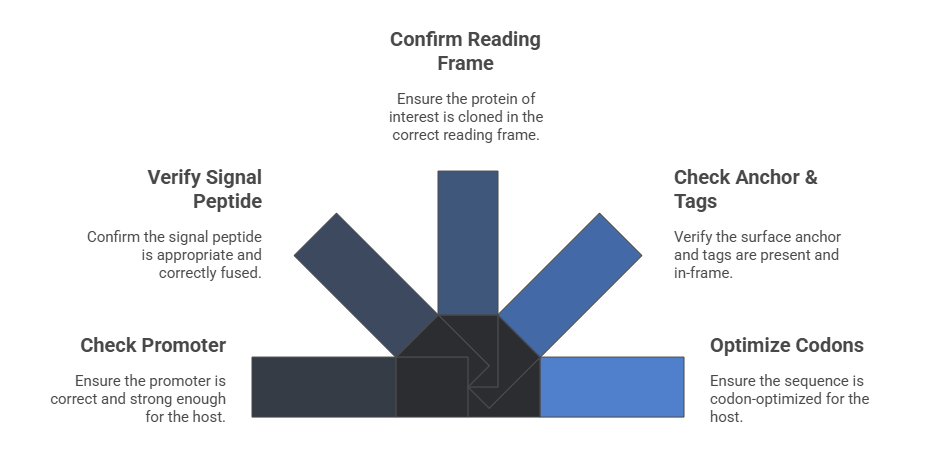
Step 1: Interrogate Your Plasmid Construct An error in the DNA is the most common cause of a global display failure. Go back to your sequence data and verify every component of your display cassette.
Promoter: Are you using the correct promoter for your host (e.g., GAL1 for yeast, CMV for mammalian)? Is it strong enough?
Signal Peptide: Is the signal peptide appropriate for your host and correctly fused in-frame to your protein? An incorrect signal peptide will prevent efficient translocation to the secretory pathway.
Protein of Interest: Confirm that your library was cloned in the correct reading frame.
Anchor & Tags: Is the surface anchor (e.g., Aga2p for yeast, a transmembrane domain for mammalian) present and in-frame? Are the expression and affinity tags correctly placed and free of stop codons?
Codon Optimization: Has the sequence been codon-optimized for your expression host? A high concentration of rare codons can stall translation and dramatically reduce protein expression.
Step 2: Scrutinize Your Host Cells The health and integrity of your host cells are non-negotiable.
Mammalian Cells:
Mycoplasma Contamination: This is a silent killer of mammalian cell experiments. Regularly test your cells, as mycoplasma can severely impact transfection efficiency and protein expression.
Cell Viability: Are your cells healthy and in the exponential growth phase before transfection? Low viability leads to poor transfection and low expression.
Yeast Cells:
Strain Integrity: Have you accidentally used the wrong yeast strain (e.g., one that is incompatible with your vector's selection marker)?
Viability: Check your yeast under a microscope. Ensure they are healthy and not overly budded or dead before induction.
Step 3: Audit Your Protocol Even with a perfect construct and healthy cells, a flawed protocol will lead to failure.
Induction (Yeast): This is the most frequent point of error. Are you inducing in a galactose-containing medium (e.g., SG-CAA) and ensuring there is absolutely no glucose, which represses the GAL1 promoter? Have you optimized the induction temperature (often 20°C is better than 30°C) and duration (16-24 hours is typical)?
Transfection (Mammalian): What is your transfection efficiency? Use a GFP-co-transfection control to quantify it. Low efficiency means only a small fraction of your cells are receiving the plasmid. Optimize your DNA quantity, reagent-to-DNA ratio, and cell density. Ensure you are harvesting cells at the optimal time post-transfection (typically 24-48 hours).
Path 2: Troubleshooting Clone-Specific Low Display (A Developability Problem)
If your system is working (i.e., control proteins display well) but specific variants show poor display, then the protein itself is the problem. This is a critical, early-warning sign of poor developability.
Step 4: Analyze the Protein Variant Itself Low display for a specific clone is often a result of the cell's own quality control machinery. If a variant is unstable or misfolds, it is targeted for degradation in the endoplasmic reticulum via ER-associated degradation (ERAD) and never reaches the cell surface.
Intrinsic Instability: The variant may have a low melting temperature (Tm) and be inherently unstable. The selection pressure might have favored mutations that improve binding at the cost of stability.
Exposed Hydrophobic Patches: Mutations can expose hydrophobic regions, leading to aggregation within the secretory pathway and triggering degradation.
Unpaired Cysteines: The introduction of an odd number of cysteine residues can lead to improper disulfide bonding, misfolding, and aggregation.
Toxicity: The protein variant itself might be toxic to the host cell, leading to reduced growth and protein synthesis.
The Solution: This is not a simple fix but rather a valuable data point. The clones with poor display are likely poor drug candidates. Prioritize clones that maintain high display levels throughout the selection process. If your best binders consistently show low display, it may be necessary to engineer them for improved stability or screen from a different library scaffold.
Conclusion: Low Display is a Feature, Not Just a Bug
While frustrating, low display levels are a critical source of information. When observed in specific clones, low display is an early and invaluable filter for poor developability, allowing you to eliminate problematic candidates long before you invest significant time and resources in their downstream characterization. By paying close attention to this metric, you can guide your engineering efforts toward variants that are not only potent but also stable and well-behaved.
De-Risk Your Biological Development Pipeline
Integrating Developability Assessment from day one is key to success. Our Ph.D level team acts as a seamless extension of yours, providing the critical data you need to select and engineer candidates with the highest probabiliy of success.
Let's discuss how we can accelerate your next project
