Accelerate your protein engineering. Download our free guide to cell display
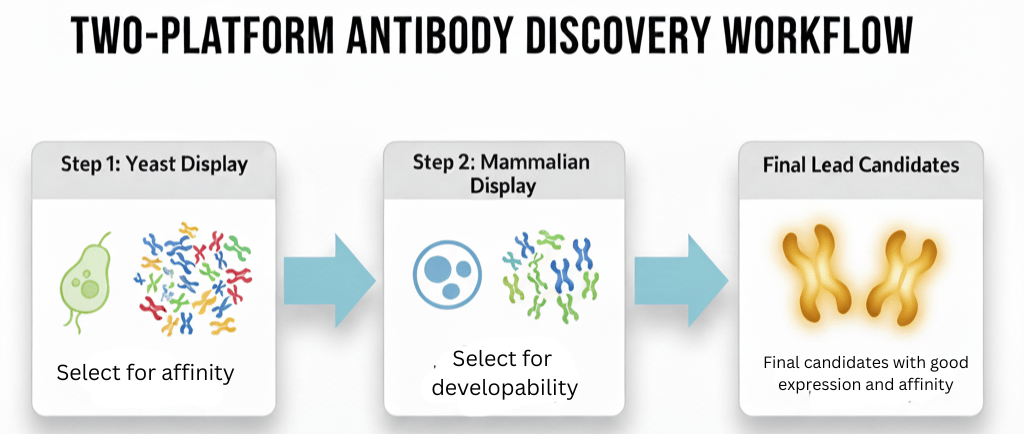
The Two-Platform Approach: Using Yeast Display for Affinity and Mammalian Display for Developability
Don't choose between yeast display and mammalian display. This guide details a two-platform biologics discovery workflow, using yeast for affinity and mammalian display to screen for developability
11/4/20253 min read
We’ve all been there.
You completed a well planned yeast display campaign. You performed incredible electroporations that resulted in a massive 10⁹ library, and after four rounds of sorting, you hit gold: a panel of binders with picomolar affinity. You pick the top candidate, spend two weeks sub-cloning it for soluble expression in a mammalian cell line, and... nothing.
The expression is terrible. The little protein you do get is 50% aggregated.
It's an incredibly frustrating and shocking moment. This single clone, which looked like a hero in yeast, failed the first real-world test. Why? Because the workflow was flawed. We asked the wrong question, and we used the wrong tool.
For years, the field has debated yeast display vs. mammalian display as if they were competitors. The truth is, they are specialized partners. Using only one is like trying to build a house with just a hammer.
Complete biologics discovery isn't about choosing one platform over the other; it's about using them in a sophisticated, two-step workflow. Use yeast for what it’s best at: raw discovery. Then, use mammalian for what it's best at: validation and developability.
Step 1: Yeast Display as the Discovery Engine
First, let's respect what yeast display brings to the table. It is, without a doubt, the most powerful discovery engine we have.
Massive Numbers: You can screen highly diverse libraries, giving you unparalleled statistical power to find incredibly rare binding events.
Robustness and Speed: Yeast is tough. It grows fast, it's easy to handle, and the display is highly stable and uniform
A "Pure" Affinity Readout: The yeast cell wall is a relatively simple environment. This makes it a fantastic, clean system for selecting one thing: pure binding affinity and specificity.
The goal of Step 1 is to harness this power. Go broad. Run your full-scale library through 3-4 rounds of high-stringency sorting in yeast. Your objective is not to find one "winner." Your objective is to find the top hits —the enriched polyclonal pool you get right before you'd normally pick single clones.
Then, you stop. You don't pick a winner. You use NGS to get the sequences of that entire enriched pool.
Step 2: Mammalian Display as the "Developability-Matrix"
This is where next-level protein engineering begins. That polyclonal pool of hits from your yeast screen is your new starting library. It's an elite library where every member is already a high-affinity binder.
Now, you ask a completely different question: "Of these 10,000 great binders, which ones can actually be manufactured?"
This is a question yeast cannot answer. It can't tell you if a protein will fold correctly in a CHO cell, if it will receive the right, human-like PTMs (like glycosylation), or if it will be stable and soluble at high concentrations. For that, you need mammalian display.
Here is the workflow:
Create a Focused "Hits Library": Synthesize a new library, not of 10⁹ random variants, but of the 1,000 or 10,000 hits you identified from your yeast screen's NGS data.
Screen in Mammalian Display: Transfect this new, elite library into a mammalian cell line (like HEK or CHO) using CRISPR integration or lentivirus transfection.
Run the "Developability Sort": This is the key. You are not sorting for affinity. You already did that. Instead, you stain the cells with an antibody against the expression tag (like c-myc or HA).
Sort for the Brightest Cells: You simply collect the top 5-10% of cells with the highest expression signal.
This simple sort is what I call "Developability-Matrixing." You are running your panel of high-affinity hits through a matrix that filters for one thing: recombinant protein expression and stability.
The Payoff: Why This Workflow is Better
A clone that shows up as "high-affinity" in yeast and "high-expression" in mammalian display is a golden candidate. You have prospectively selected for a molecule that has passed the two most important tests:
It has high function (from yeast).
It has high developability (from mammalian)
This two-platform approach "shifts left" on developability, pulling a critical, late-stage failure point all the way to the front of the discovery process. You are no longer guessing which yeast hit will work. You are generating a final, prioritized list of candidates that are already proven to be well-behaved in the very system you will use to manufacture them.
Implementing a sophisticated two-platform screening strategy requires careful design and execution, from NGS-guided library synthesis to high-throughput mammalian cell screening. To learn how Ranomics' expert protein engineering services can help you de-risk your biologics discovery pipeline and identify high-quality, developable leads, contact our team today
Get in touch
Looking to perform yeast display screening or mammalian display screening.
Get in touch with our expert team today
