Accelerate your protein engineering. Download our free guide to cell display
Introduction to Protein Developability: What Makes a Good Biologic Drug?
A biologic with high potency is only half the battle, as many promising candidates fail due to poor developability and manufacturability. This guide introduces the core pillars of a successful biologic, including robust expression, biophysical stability, high solubility, and the avoidance of chemical liabilities. Integrating these developability assessments early in the discovery process is critical to de-risking drug development and creating molecules that can become effective medicines.
9/29/20253 min read
In the world of biologics development, a successful discovery campaign will end with the identification of a lead candidate with exquisite potency and specificity. The biologic will bind its target with high affinity and specificity and also potently activate or block targeted biological events. However, this is only half the battle. The path from a promising lab-scale hit to an approved FDA drug is fraught with challenges, and the vast majority of candidates fail, not because they lack efficacy, but because they possess poor developability or manufacturability.
Developability and manufacturability refers to the collection of biophysical and biochemical properties that determine whether a biologic can be manufactured at high yield, formulated at high concentration, and remain stable and safe until administration to a patient.
Historically, developability and manufacturability was assessed late in the development cycle, leading to costly failures. The modern way of thinking is to integrate this assessment into the earliest stages of discovery. This post provides an introduction to the core pillars of protein developability and why they are essential for creating a successful biologic.
Pillar 1: Manufacturability and Expression
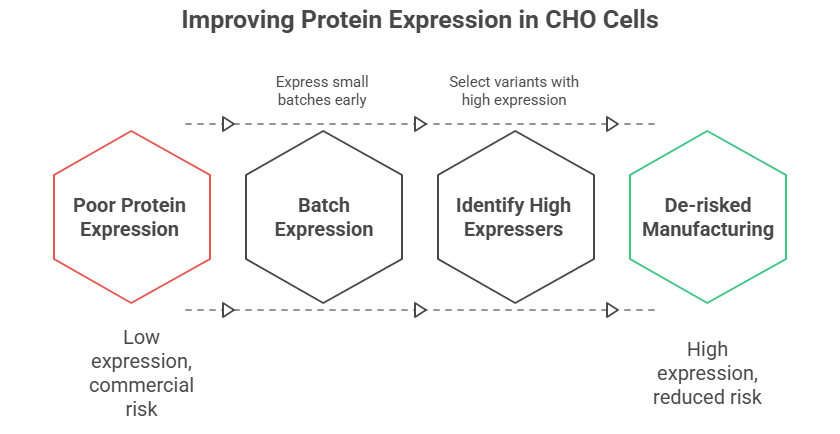
A biologic drug is useless if it cannot be produced at a commercial scale. The gold standard for therapeutic antibody production is expression in Chinese Hamster Ovary (CHO) cells, and a key metric is the expression titer, measured in grams per liter (g/L).
The Challenge: A candidate that expresses poorly (e.g., <1 g/L) in a standard CHO line presents a significant commercial risk. Poor expression can be caused by a variety of factors, including inefficient transcription, poor protein folding and trafficking, or cellular toxicity.
Early Assessment: During lead selection, expressing small batches of top candidates in a representative host system provides an early indication of their manufacturing potential. Variants with inherently high expression profiles are significantly de-risked.
Pillar 2: Stability (Thermodynamic and Colloidal)
A successful biologic must be stable enough to withstand the stresses of purification, formulation, long-term storage, and administration. Stability is broadly divided into two categories.
Thermodynamic Stability: This is a measure of a protein's resistance to unfolding, often quantified by its melting temperature (Tm). A higher Tm indicates a more stable protein that is less likely to denature. A low Tm can lead to aggregation and loss of function.
Colloidal Stability: This describes the tendency of protein molecules to interact with each other at high concentrations. Poor colloidal stability leads to high viscosity and aggregation, making the drug difficult to administer (especially for subcutaneous injections) and potentially immunogenic.
Key Assays: Techniques like Differential Scanning Calorimetry (DSC) or Differential Scanning Fluorimetry (DSF) are used to measure Tm, while Dynamic Light Scattering (DLS) and Self-Interaction Nanoparticle Spectroscopy (SINS) can predict colloidal stability.
Pillar 3: High Solubility and Low Aggregation Propensity
Aggregation is the irreversible self-association of protein molecules and is one of the primary failure modes for biologics. It can lead to loss of efficacy and, more dangerously, can trigger a severe immunogenic response in patients.
The Challenge: Aggregation is driven by the exposure of hydrophobic or charged patches on the protein surface. Many modern biologic formulations require extremely high concentrations (>100 mg/mL), which dramatically increases the risk of aggregation.
Early Assessment: In silico tools can predict aggregation-prone regions by analyzing a protein's surface hydrophobicity. Experimentally, techniques like Size Exclusion Chromatography (SEC) are used to detect the presence of soluble aggregates, while accelerated stability studies (involving thermal or chemical stress) can reveal a candidate's long-term aggregation propensity.
Pillar 4: Chemical Stability and Lack of Post-Translational "Hotspots"
Over time, proteins can undergo chemical degradation, leading to heterogeneity and loss of function. A good biologic should be free of common chemical liability "hotspots."
Common Liabilities:
Deamidation: The conversion of asparagine (N) to aspartic acid, especially in N-G (Asn-Gly) motifs.
Isomerization: The conversion of aspartic acid (D) to its stereoisomer, isoaspartate.
Oxidation: The modification of methionine (M) and tryptophan (W) residues.
Unpaired Cysteines: Free thiol groups that can lead to incorrect disulfide bond formation and aggregation.
Glycosylation Sites: Unintended N-linked glycosylation sites (N-X-S/T motifs) can introduce unwanted heterogeneity.
Early Assessment: These hotspots can be identified and flagged in silico directly from a variant's amino acid sequence. During candidate selection, variants free of these liabilities should be prioritized.
Conclusion: Integrating Developability from Day One
A successful biologic drug is a finely tuned balance of potent biological function and robust, drug-like properties. By treating developability not as a late-stage hurdle but as a critical part of the initial discovery process, we can significantly de-risk drug development pipelines. Analyzing NGS data for sequence liabilities, performing early-stage biophysical characterization, and prioritizing candidates with favorable stability and solubility profiles are essential steps. This "front-loading" of developability assessment ensures that the immense effort of a discovery campaign is directed toward creating molecules that have
De-Risk Your Biological Development Pipeline
Integrating developability assessment from day one is key to success. Our Ph.D.-level team acts as a seamless extension of yours, providing the critical data you need to select and engineer candidates with the highest probability of success.
Let's discuss how we can accelerate your next project.
