Accelerate your protein engineering. Download our free guide to cell display
Eliminating False Positives: Mastering Avidity Effects in Yeast Display Screening
Avidity effects in yeast and mammalian display screening create false positives that waste time and resources in drug development. This guide provides strategies for identifying and eliminating these artifacts through advanced screening methodologies, helping research teams avoid costly mistakes and improve therapeutic protein development success rates.
8/11/20256 min read
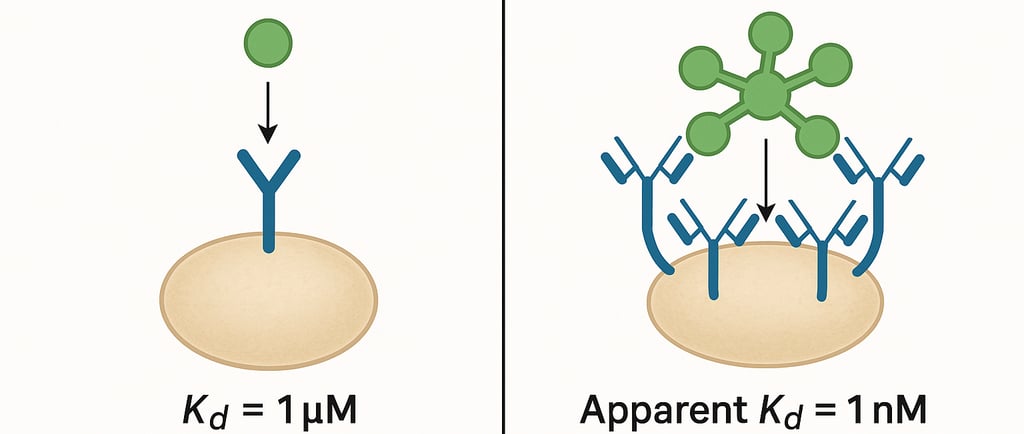
One common challenge facing protein engineers using yeast display and mammalian display technologies is the phenomenon of avidity effects. Avidity effects can lead to the selection of false positive variants that appear to exhibit high-affinity binding during screening but fail to perform when produced as soluble proteins. For biotechnology and pharmaceutical companies investing significant resources in therapeutic protein development, understanding and mitigating these effects is crucial for ensuring that screening campaigns identify genuinely valuable candidates rather than false positive artifacts of the display system.
Understanding Avidity: The Multi-valency Problem
Avidity refers to the combined strength of multiple binding interactions between multivalent molecules. In yeast display and mammalian display systems, each cell presents thousands of copies of the target protein on its surface. Multiple copies create a high local concentration that can lead to dramatically enhanced apparent binding affinity compared to the intrinsic monovalent affinity of individual protein-target pairs. This enhancement can be several orders of magnitude, making proteins with relatively weak intrinsic affinities appear to be high-affinity binders during screening. The density of displayed proteins on the cell surface, typically 10^4 to 10^5 copies per cell, creates multiple opportunities for simultaneous binding events that synergistically enhance the overall binding strength.
The geometry and valency of the target molecule play important roles in determining the magnitude of avidity effects. Cell surface receptors that exist as higher-order oligomers, viral particles presenting multiple copies of surface proteins, and protein aggregates containing repeated epitopes all provide multiple binding sites that can be simultaneously engaged by displayed proteins. The spatial arrangement of these binding sites and the flexibility of the linkages connecting displayed proteins to the cell surface determine whether simultaneous binding events are geometrically feasible.
This phenomenon is particularly problematic for developing therapeutic antibodies, where the ultimate application requires monovalent binding interactions. Antibody fragments that demonstrate strong binding to cell-surface antigens during yeast display screening may exhibit disappointingly weak binding when produced as soluble proteins and tested in conventional binding assays. This discrepancy can lead to significant waste of time and resources as development teams pursue promising leads that ultimately fail to meet performance criteria in downstream applications.
The temporal aspects of avidity effects add another layer of complexity to the challenge. Polyvalent interactions typically exhibit slower dissociation kinetics than their monovalent counterparts, meaning that standard washing procedures during flow cytometry may not effectively remove weakly bound targets from cells displaying low-affinity proteins. This can result in the apparent enrichment of weak binders during selection campaigns, particularly when using short incubation and washing times that are insufficient to reach equilibrium.
Kinetic Screening Strategies for Authentic Affinity Assessment
The most powerful approach for mitigating avidity effects involves the implementation of kinetic screening strategies that focus on the measurement of dissociation rate constants (koff) rather than equilibrium binding constants. This approach recognizes that while avidity effects can dramatically enhance apparent equilibrium affinity, they have a much smaller impact on the intrinsic dissociation rate constants of individual binding events.
Kinetic screening protocols typically begin with the incubation of yeast cells with fluorescently labeled target molecules under equilibrium conditions, allowing sufficient time for binding interactions to reach steady state. After washing to remove unbound target molecules, the cells are resuspended in a solution containing a large excess of unlabeled competitor molecules that prevent the rebinding of dissociated labeled targets. The rate of fluorescence loss from the cell population is then monitored over time, providing a direct measurement of dissociation kinetics.
The success of kinetic screening strategies depends critically on the selection and optimization of competitor molecules and experimental conditions. The competitor should possess sufficient affinity for the target to effectively prevent rebinding while not interfering with the measurement of dissociation from displayed proteins . In many applications, the unlabeled version of the target molecule itself serves as an effective competitor, but alternative competitors such as related proteins or small molecule inhibitors may be more appropriate for specific targets or applications.
The concentration of competitor molecules must be carefully optimized to ensure effective competition without introducing artifacts or complications. Typically, competitor concentrations should be 10-100 fold higher than the target concentration to ensure effective prevention of rebinding. However, excessively high competitor concentrations may lead to non-specific effects or interference with the measurement system, requiring careful optimization for each specific application.
Off-Rate Selection: Precision Engineering for High-Affinity Variants
Off-rate selection represents a specialized form of kinetic screening that has proven particularly effective for identifying high-affinity antibody variants and other binding proteins. This approach involves the incubation of yeast libraries with labeled antigens followed by extensive washing under conditions that promotes the dissociation of weakly bound complexes.
The implementation of off-rate selection requires careful optimization of washing conditions to achieve the desired selection pressure. The duration of washing steps must be sufficient to allow weakly bound variants to dissociate while preserving strongly bound variants. The use of multiple washing steps with increasing stringency can provide fine control over the selection pressure and enable the identification of variants with exceptional binding properties.
Buffer composition during off-rate selection can significantly influence the effectiveness of the selection process. The inclusion of chaotropic agents can help destabilize weak interactions while preserving strong ones. However, the concentration of these agents must be carefully optimized to avoid disrupting even strong binding interactions or causing protein denaturation.
The pH and ionic strength of washing buffers can also be manipulated to create selective pressure for variants with specific binding properties. Careful consideration must be given to ensure that buffer conditions do not deviate too far from physiologically relevant conditions.
Competitive Binding Assays: Validating Specificity and Affinity
Competitive binding assays provide another powerful tool for distinguishing between genuine high-affinity interactions and avidity artifacts. These assays involve the simultaneous incubation of yeast-displayed proteins with both labeled targets and competing molecules, allowing for the assessment of binding specificity and the identification of variants that maintain high affinity even in the presence of competitors.
The design of effective competitive binding assays requires careful consideration of competitor selection and concentration optimization. Competitors should be chosen to challenge specific aspects of the binding interaction, such as overall affinity, binding specificity, or resistance to interference from related molecules. The use of multiple competitors with different binding modes can provide comprehensive information about the binding mechanism and help identify variants with unique or superior binding properties.
Concentration-response curves using different competitor concentrations can provide quantitative information about binding affinity and help establish structure-activity relationships for displayed variants. These curves can be analyzed using standard pharmacological methods to determine IC50 values and other quantitative measures of binding affinity. The comparison of IC50 values obtained in competitive binding assays with apparent affinities measured in direct binding assays can provide insights into the extent of avidity effects.
The temporal aspects of competitive binding assays must be carefully controlled to ensure meaningful results. Sufficient incubation time must be allowed for binding interactions to reach equilibrium, while avoiding conditions that might lead to protein degradation or other artifacts. The use of kinetic analysis methods can help determine optimal incubation times and provide additional information about binding mechanisms.
Advanced Flow Cytometry Techniques
Modern flow cytometry instruments offer sophisticated capabilities that can be leveraged to improve the detection and mitigation of avidity effects in yeast display screening. Multi-parameter analysis enables the simultaneous measurement of multiple fluorescent signals, allowing for the assessment of protein expression levels, binding activity, and other relevant parameters in single experiments.
The use of ratiometric analysis, where binding signals are normalized to expression levels, can help identify and correct for expression-level bias that might confound the interpretation of binding results. This approach is particularly valuable when screening libraries with heterogeneous expression levels or when comparing variants with different expression characteristics.
Time-resolved flow cytometry measurements can provide kinetic information about binding and dissociation processes, enabling the identification of variants with specific kinetic properties. These measurements can be particularly valuable for off-rate selection experiments and for validating the results of kinetic screening assays.
Expert Solutions for Avidity-Free Screening
Navigating the complexities of avidity effects requires deep expertise in yeast display and mammalian display technology and extensive experience with advanced screening methodologies. Ranomics brings this expertise to biotech and pharmaceutical companies through comprehensive screening services that incorporate state-of-the-art approaches for eliminating false positives and identifying genuinely high-affinity variants.
Our team has developed proprietary screening protocols that combine different analyses and validation to ensure that selected variants will perform well in downstream applications. We understand that avidity artifacts can be particularly costly for pharmaceutical companies, where false positives can lead to expensive development programs that ultimately fail to deliver viable therapeutic candidates.
Our experience with both yeast display and mammalian display systems enables us to select the optimal platform for each specific application and to implement appropriate controls and validation strategies.
The challenge of avidity effects is not merely technical but also strategic, requiring careful consideration of screening objectives, validation requirements, and downstream application needs. Our team works closely with clients to develop customized screening strategies that balance the need for high-throughput identification of promising variants with the requirement for rigorous validation to eliminate false positives.
For biotech and pharmaceutical companies seeking to maximize the value of their protein engineering investments, partnering with Ranomics provides access to the expertise and methodologies needed to overcome avidity challenges and identify genuinely valuable therapeutic candidates. Our commitment to staying at the forefront of screening technology ensures that our clients benefit from the latest advances in kinetic analysis, competitive binding assays, and validation methodologies.
Whether you're developing therapeutic antibodies, optimizing binding proteins, or engineering novel molecular recognition systems, Ranomics has the expertise and capabilities to help you navigate the complexities of avidity effects and achieve your protein engineering objectives efficiently and cost-effectively.
